Marginal effects: getting insights from a model
2023-04-28
Introduction
Learning objective
Using R and/or SAS:
- Understand limitation of typical regression output;
- Learn how to estimate marginal effects for categorical/continuous variables;
- Learn how to visualize regression models;
- Learn how to estimate custom hypotheses.
Standard regression output
Regression models summarize data, but the standard output of a regression analysis is not automatically informative.
Model (estimator) \(\text{HorsePower}_i=\beta_0+\beta_X\text{Cylinder}_i+\epsilon_i\)
Typical R output
Call: glm(formula = hp ~ cyl, data = mtcars)
Coefficients:
(Intercept) cyl
-51.05 31.96
Degrees of Freedom: 31 Total (i.e. Null); 30 Residual
Null Deviance: 145700
Residual Deviance: 44740 AIC: 328.6Typical SAS output
Definition
Marginal effects can help interpreting regression models (Norton, Dowd, and Maciejewski 2019).
- Informally, a marginal effect is describing the change in outcome \(Y\) given \(X\), but according to population average of the other covariates.
- In other words, making your model (more) interpretable
- Note that the terminology is extremely confusing
Issue
For simple models, the marginal effect is straightforward.
- The marginal effect in \(Y=\beta_0+\beta_XX+\epsilon\) is…
- \(\beta_X\) !
For more complex models, marginal effects are harder to obtain
- \(\text{logit}(Y)=\beta_0+\beta_X\text{rcs}_X+\beta_{X_2}*\beta_{X_3}+\gamma Z\)
Package to derive marginal effects
R packages were developed to facilitate marginal effect estimation
marginaleffects(Arel-Bundock 2023)emmeans(includes the olderlsmeans)
Equivalent SAS options:
ESTIMATELSMEANS
Example data
Demonstration data
Data from the Canadian Community Health Survey (CCHS) 2015 - Nutrition is used.
- Survey design and sampling weights are not considered
- Assume the sample is just any random sample of the population
- Proper survey analysis is described elsewhere and on my blog (shameless promotion)
Load demonstration data
Code
# Loading
load(file = here::here("data","processed","cchs2015_demonstration.rdata"))
# Select only some variables
final <-
cchs2015_demonstration |>
dplyr::select(participantid, r24_weekend, age, sex, education,
smoking, phys_act_mod, self_reported_bp,
energy, sodium, fibers)
# preview
head(final,3)# A tibble: 3 × 11
participantid r24_weekend age sex education smoking phys_act_mod
<dbl> <dbl> <dbl> <fct> <dbl> <dbl> <dbl>
1 9261 1 20 Male 2 0 4.5
2 1881 0 56 Male 3 0 5.25
3 16071 1 35 Female 4 0 2.25
# ℹ 4 more variables: self_reported_bp <dbl>, energy <dbl>, sodium <dbl>,
# fibers <dbl>Example application
Descriptive analysis: E(energy | age, sex, physical activity)
For descriptive purpose, we wish to estimate self-reported energy intakes according to age (years), sex and moderate/vigorous physical activity (hours/week).
Model (estimator) \(\text{Energy}_i=\beta_0 + \beta_1\text{age} + \beta_2\text{female} + \beta_3\text{phys. act.} +\epsilon_i\)
SAS equivalent:
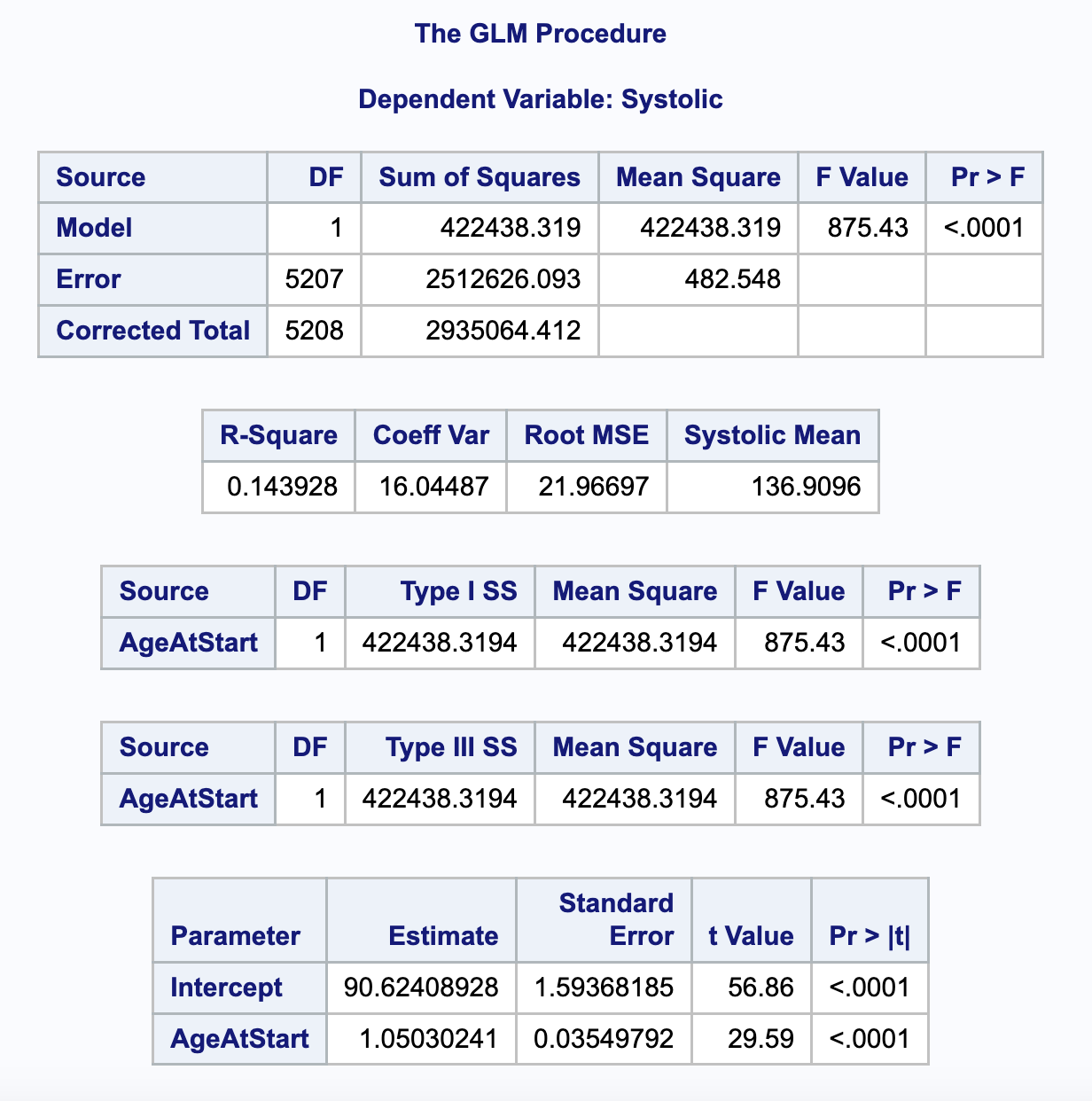
Model summary
What can we say based on this output?
Call: glm(formula = energy ~ age + sex + phys_act_mod, family = gaussian(link = "identity"),
data = final)
Coefficients:
(Intercept) age sexFemale phys_act_mod
2492.535 -7.477 -539.742 21.046
Degrees of Freedom: 997 Total (i.e. Null); 994 Residual
(2 observations deleted due to missingness)
Null Deviance: 835200000
Residual Deviance: 736100000 AIC: 16330Exploring parameters: overview
Helpful package to assess coefficients and generate standard output:
broom::tidy(model)parameters::parameters(model)gtsummary::tbl_regression()
SAS equivalent: ods output <...>; (see procedure documentation or use ods trace on;)
Exploring parameters: example
Model assumptions: overview
Linear regression models have 4 key assumptions.
- Independence;
- Homoscedasticity: to verify;
- Normality: to verify;
- Linearity: to verify.
Model assumptions: example
To calculate model errors (residuals): residuals(lm1)
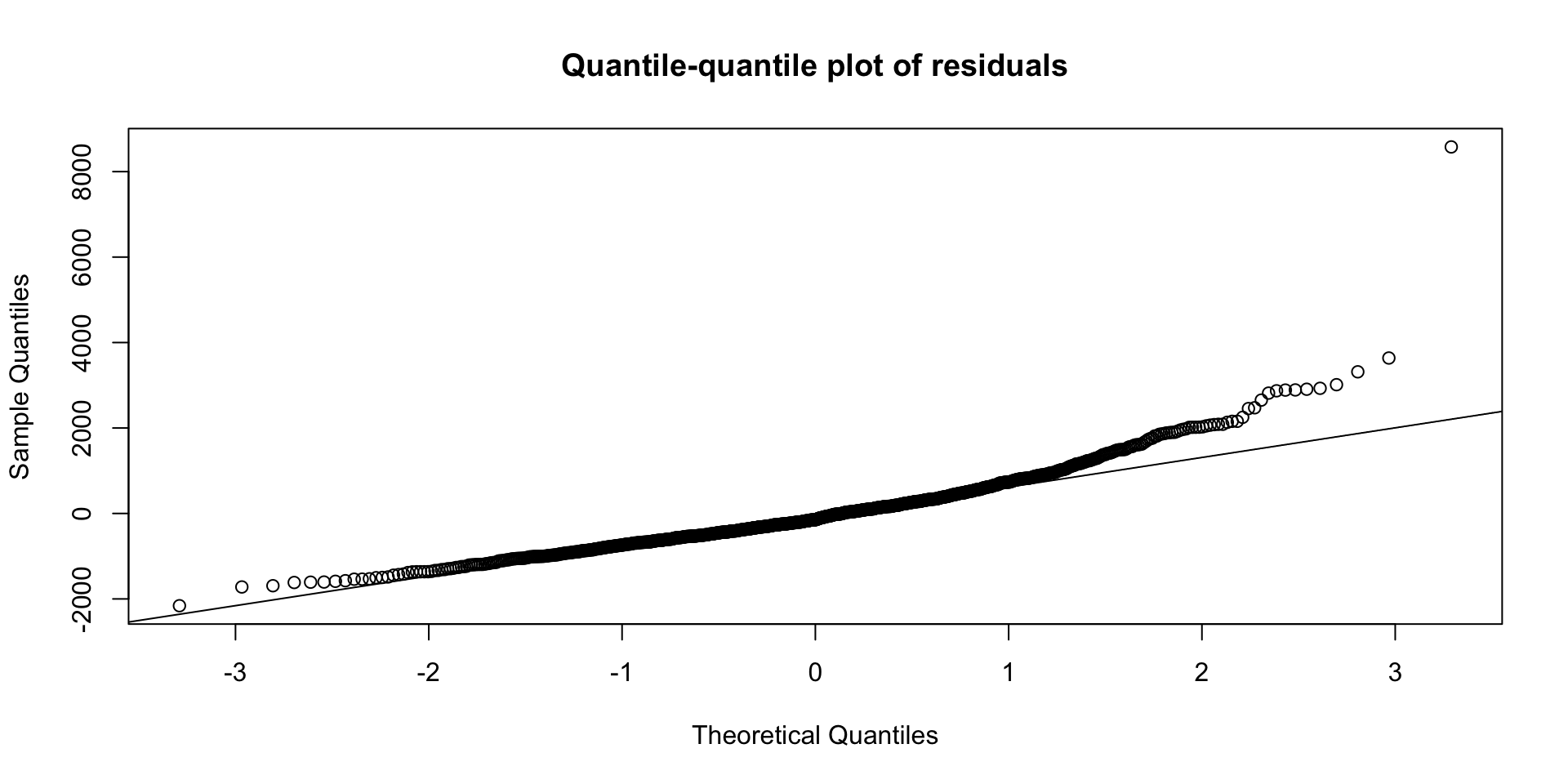
What can we say about this graph?
Model fine-tuning
- Independence: assumed by design;
- Homoscedasticity: log transformation of energy
- Normality: log transformation of energy
- Linearity: restricted cubic spline transformation
Of note, log transformation not shown here for simplicity.
Revised model
Specify Restricted Cubic Spline (RCS) transformation with the rms package.
rms::rcs(variable,knots)
Exploring parameters: example
| Characteristic | Beta | 95% CI1 | p-value |
|---|---|---|---|
| Age, years | |||
| rms::rcs(age, 4)age | -16 | -34, 1.0 | 0.065 |
| rms::rcs(age, 4)age' | 21 | -26, 69 | 0.4 |
| rms::rcs(age, 4)age'' | -65 | -234, 104 | 0.5 |
| sex | |||
| Male | — | — | |
| Female | -545 | -653, -436 | <0.001 |
| Moderate or vigorous physical activity, hours/week | |||
| rms::rcs(phys_act_mod, 3)phys_act_mod | -13 | -61, 36 | 0.6 |
| rms::rcs(phys_act_mod, 3)phys_act_mod' | 90 | -34, 213 | 0.2 |
| 1 CI = Confidence Interval | |||
… How can we interpret this revised model?
Categorical covariate/group mean (1)
Marginal Means/Least-square means: “Predictions of a model, averaged across a ‘reference grid’ of categorical predictors.”
Categorical covariate/group mean (2)
More flexible analysis with marginaleffects
Term Value Mean Pr(>|z|) 2.5 % 97.5 %
sex Male 2157 <0.001 2040 2274
sex Female 1612 <0.001 1493 1732
Results averaged over levels of: sex
Columns: rowid, term, value, sex, estimate, p.value, conf.low, conf.high, age, energy, phys_act_mod, wts Categorical covariate/group mean (SAS)
Least-Square Means: LS-means are predicted population margins—that is, they estimate the marginal means over a balanced population.
SAS equivalent: LSMEANS sex ;
Categorical covariate/group mean difference
Term Mean Pr(>|z|) 2.5 % 97.5 %
Male - Female 545 <0.001 436 653
Results averaged over levels of: sex
Columns: term, estimate, p.value, conf.low, conf.high SAS equivalent: LSMEANS sex / diff=all cl ;
Model visualization at representative values
Plot specific parameters with marginaleffects::plot_predictions
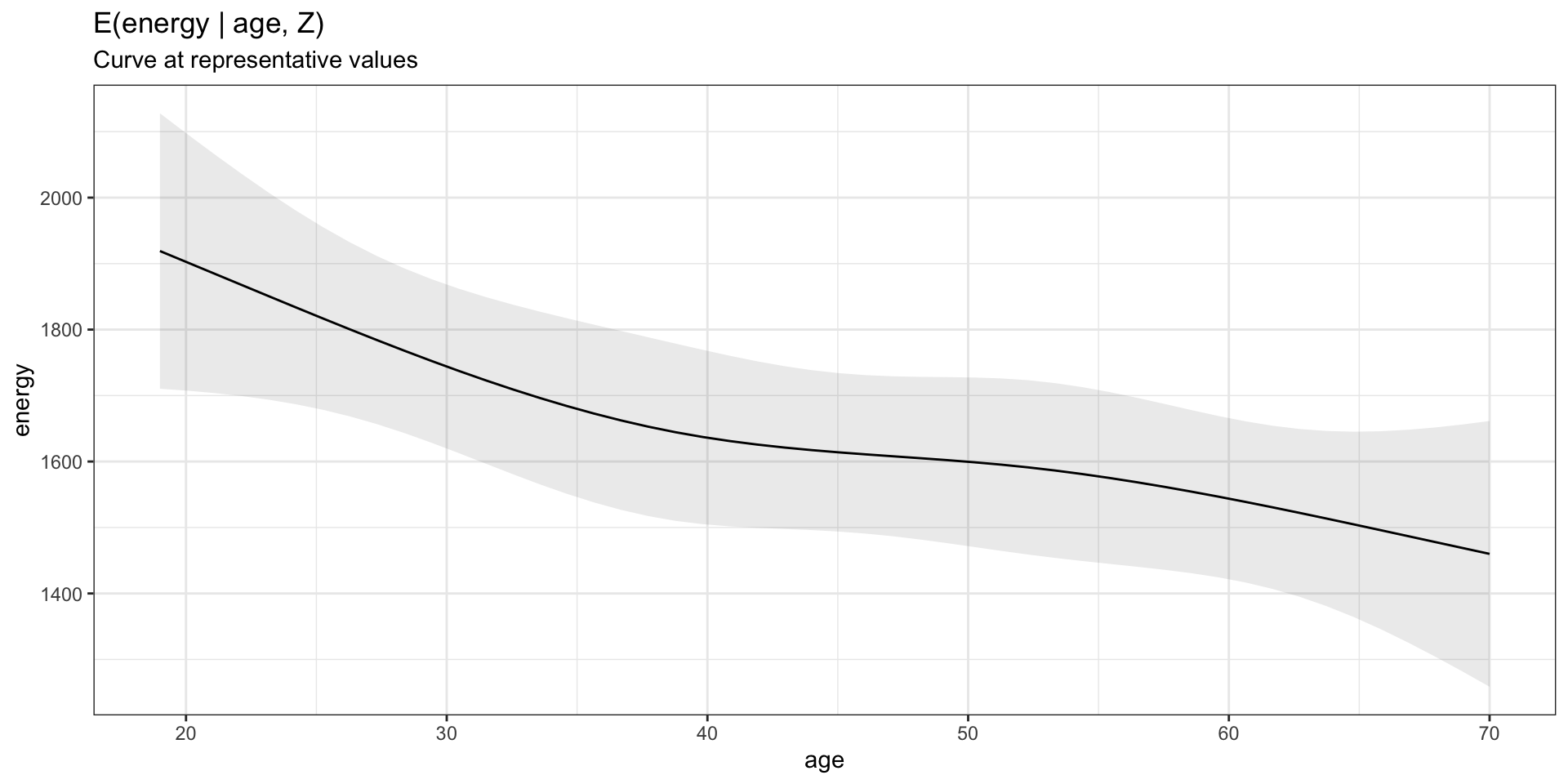
Model visualization at representative values (SAS)
SAS equivalent: proc plm ...;
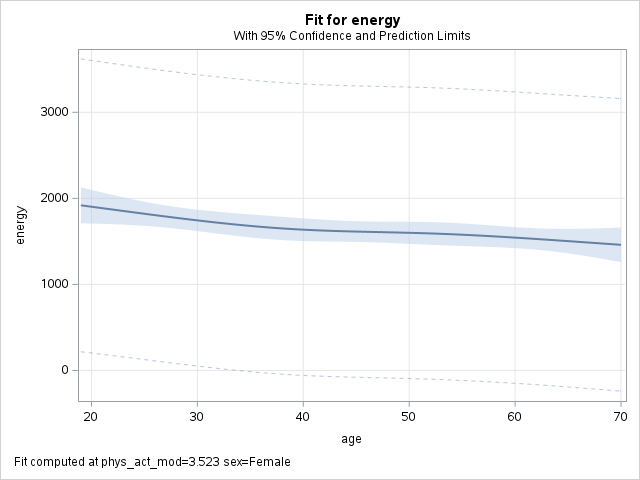
Model visualization at mean values
Alternative to representative values: use mean of all covariates, including categorical ones (coded as dummy covariates).
- Age:
mean(final$age)= 45.53 - Sex:
mean(final$female)= 0.5 - Physical act.:
mean(final$phys_act_mod)= 3.52
Model visualization at mean values
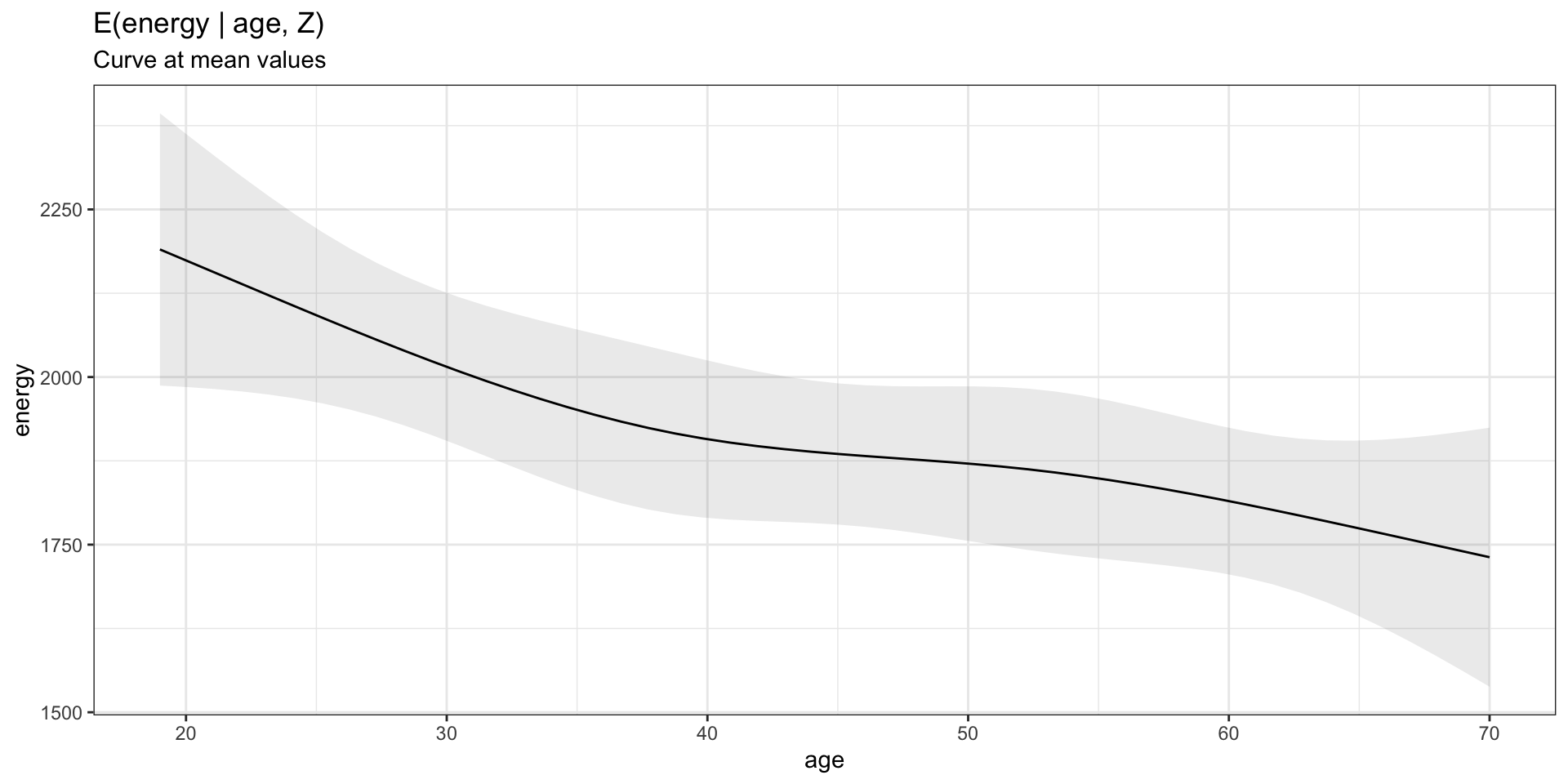
Model visualization for more than 1 covariate
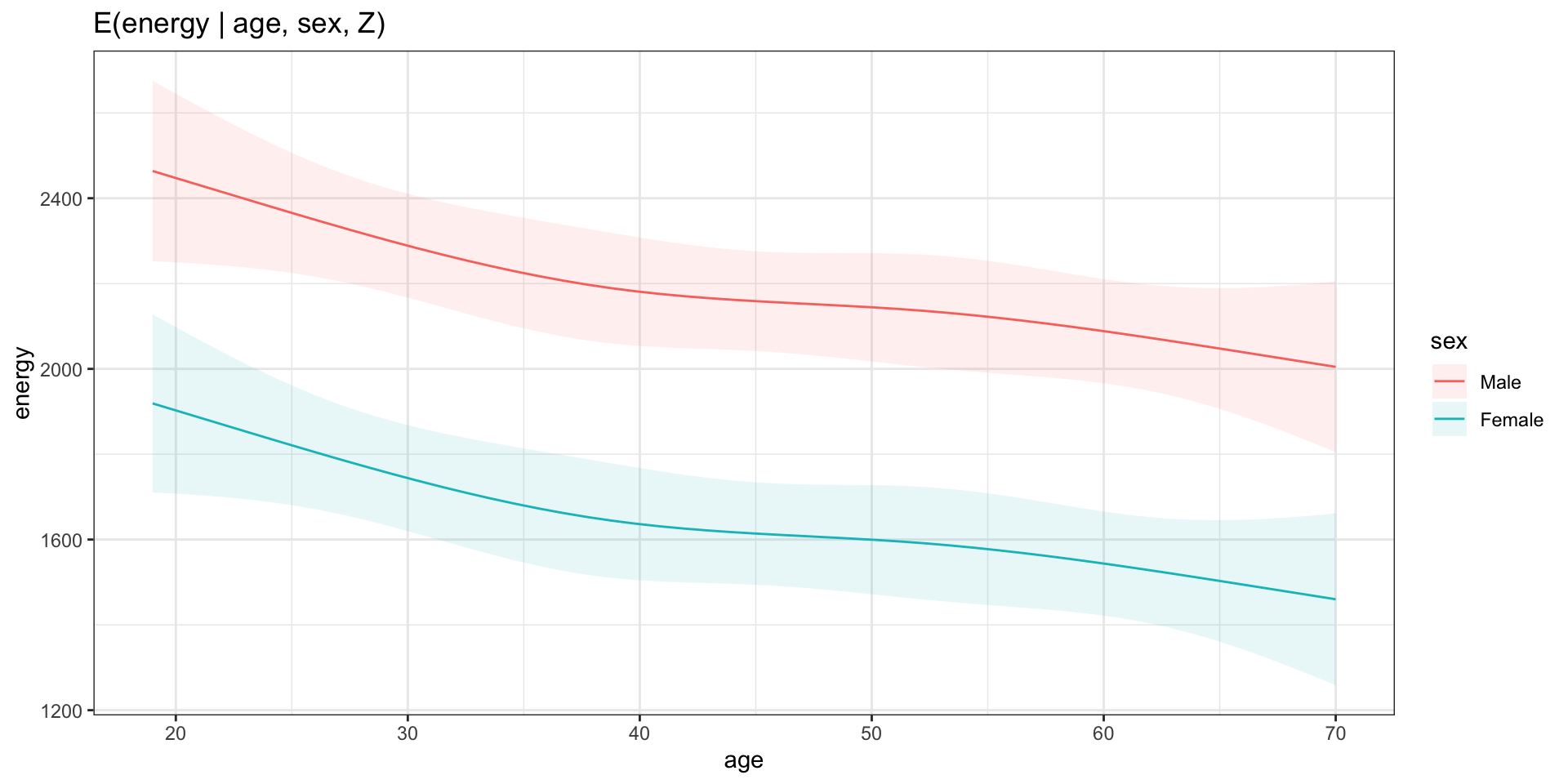
Custom hypothesis at user-defined values
Use marginaleffects::avg_comparisons
E(energy|age=60, Z) - E(energy|age=30, Z)
Code
Term Contrast Estimate Std. Error z Pr(>|z|) 2.5 % 97.5 %
age 60 - 30 -200 68.9 -2.91 0.00365 -335 -65.2
Columns: term, contrast, estimate, std.error, statistic, p.value, conf.low, conf.high Custom hypothesis for common change
E(energy|age=75th, Z) - E(energy|age=25th, Z)
Code
Term Contrast Estimate Std. Error z Pr(>|z|) 2.5 % 97.5 %
age Q3 - Q1 -126 78.2 -1.61 0.107 -279 27.3
Columns: term, contrast, estimate, std.error, statistic, p.value, conf.low, conf.high Example 2: more complex model
For causal inference purpose, we wish to estimate the risk of hypertension across the range of sodium intakes.
Model (estimator): \(\text{logit}(\pi)=\beta_0+\beta_\text{Na}\text{Na}+\gamma Z\)
Hypothesized relationship
Assume temporality and that this causal directed acyclic graph (DAG) is correct:
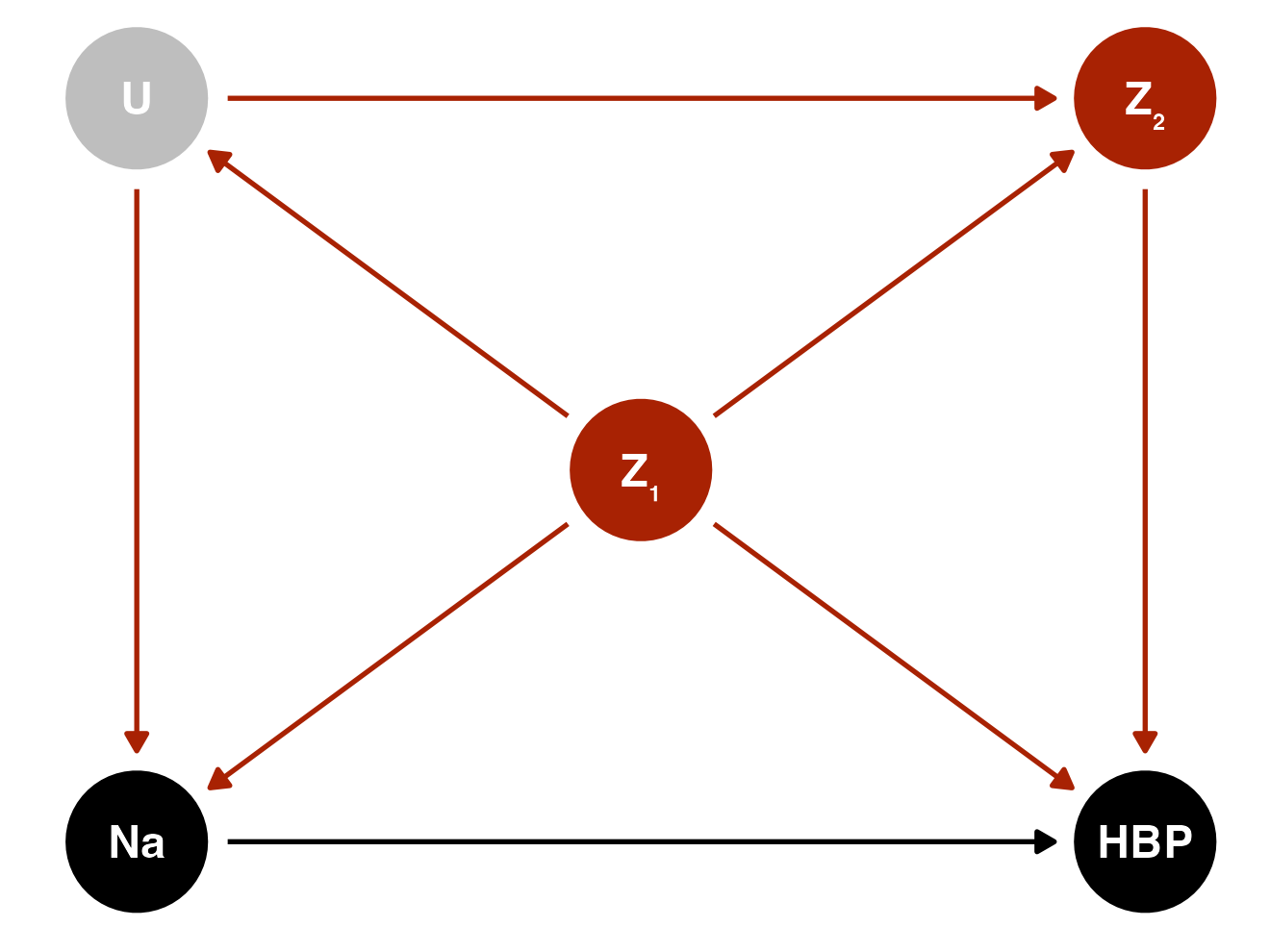
- \(\text{Na}\) sodium intake (mg)
- \(\text{HBP}\) High blood pressure
- \(U\) (unobserved) propensity towards health-seeking behaviors
- \(Z_1\) age, sex, university education level
- \(Z_2\) smoking, moderate/intense physical activity
Prepare data
Combine variables to avoid over-fitting, i.e., limiting sample size = 142 events (Harrell 2015).
Code
education
edu_4 1 2 3 4
0 84 258 348 0
1 0 0 0 300 smoking
smk_1 0 1 2
0 802 0 0
1 0 50 148Logistic regression model
Code
Call: glm(formula = self_reported_bp ~ rms::rcs(sodium, 3) + rms::rcs(age,
3) + female + edu_4 + smk_1 + rms::rcs(phys_act_mod, 3),
family = binomial(link = "logit"), data = final)
Coefficients:
(Intercept) rms::rcs(sodium, 3)sodium
-9.7611025 0.0004064
rms::rcs(sodium, 3)sodium' rms::rcs(age, 3)age
-0.0006629 0.1621546
rms::rcs(age, 3)age' female
-0.0635853 -0.3261307
edu_4 smk_1
-0.3632489 -0.3183342
rms::rcs(phys_act_mod, 3)phys_act_mod rms::rcs(phys_act_mod, 3)phys_act_mod'
0.0329085 -0.0477153
Degrees of Freedom: 986 Total (i.e. Null); 977 Residual
(13 observations deleted due to missingness)
Null Deviance: 798.7
Residual Deviance: 642.3 AIC: 662.3:::
Model visualization at mean values
Code
marginaleffects::plot_predictions(model = logm1, condition =c("sodium"),
draw=TRUE) +
# Limit axis to remove extreme values
ggplot2::coord_cartesian(xlim=c(quantile(final$sodium,0.01),
quantile(final$sodium,0.99))) +
# Add labels
ggplot2::labs(title = "Pr(High blood pressure | sodium, Z)",
subtitle = "Curve at mean values",
x = "Sodium intake, mg",
y = "Probability of high blood pressure") +
ggplot2::theme_bw() 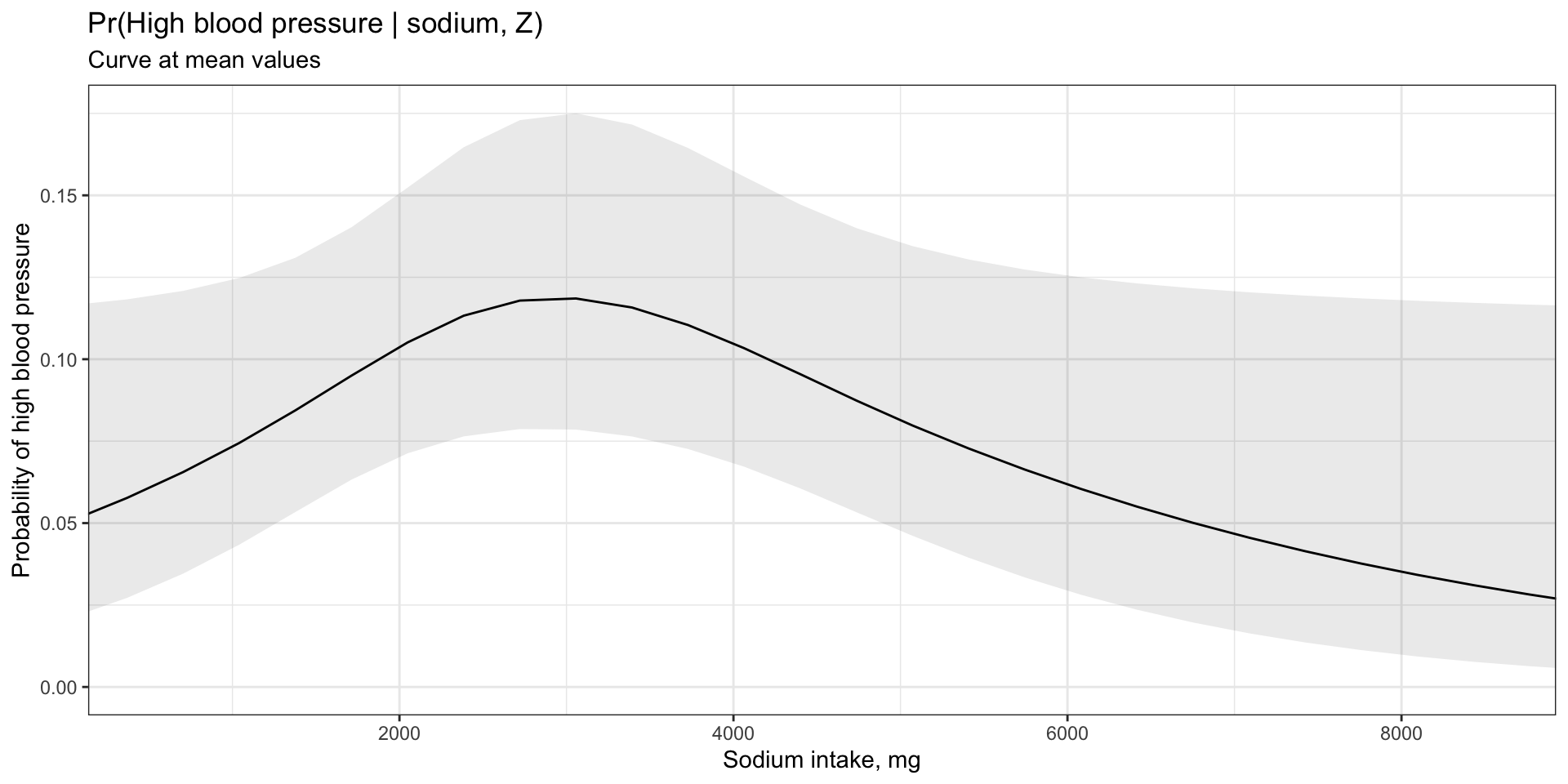
Conclusion
- Don’t stop at standard output regression model
- Marginal effects facilitate communication
- Create graph and/or test custom hypothesis
- R:
marginaleffects+ggplot2 - SAS: option
store+proc plm+proc sgplot - SAS: option
estimate+ods output <...>;
- R:
References
SHN Coding Club